MalarImDB is equipped with several search engines, for e.g. particular molecules, parasite species, treatment, text terms, etc. The different search engines can be used to retrieve literature data from the database in a simple and rapid way, thereby allowing the comparison between multiple studies with different mouse models, in different patient groups or between different malaria complications. Below some basics about the use of MalarImDB is explained, for more detailed and in-depth information, please consult our recent review (Deroost et al., Trends Parasitol. 2014, in press).
1. Each database screen contains three parts
The left part contains the search engines (see below). In the right column it is possible to select or deselect the dynamic fields, i.e. the columns you want to view in the table. When something is selected in these two parts, always press on the ENTER-key or click on the ‘filter’ button to load the data selection. To go back to the default settings or to make a new selection click on the ‘reset’ button.
The output table containing the data appears at the bottom of the page. Clicking on the arrow in the first field column of the output table retrieves a detailed view of that entry. In the last field column ‘PubMed’ the hyperlink redirects to the original paper in the PubMed database (U.S. National Library of Medicine, National Institutes of Health). The output table adjusted to the selections made with the search engines and with the chosen dynamic field selection can be exported as a csv file (scroll down to the bottom of the page and click on the ‘CSV’ button) that can be opened with standard software programs e.g. Microsoft Excel.
Figure: Database screen. This figure is also published in Deroost et al., Trends Parasitol. 2014, in press.
2. Search the database through pre-made search engines.
In the different datasets it is possible to search on molecular group, parasite strain, and a specific molecule or cell type. Depending on the type of data, each section of the database also has specific search engines. For example, to search for molecules showing higher levels in serum of patients with cerebral pathology compared to patients with severe malaria but without cerebral complications, one can select ‘serum’ in the organ search engine, and ‘>’ in the search engine for CM vs NCSM. This search can be made in one step or in multiple subsequent steps. Furthermore, multiple parameters can be selected in a specific search engine at the same time. For example, if data on ‘Plasmodium falciparum’ and ‘Plasmodium vivax’ infections are wanted, both options can be selected in the Plasmodium strain search engine by pressing the SHIFT-key and selecting the different options with the mouse.
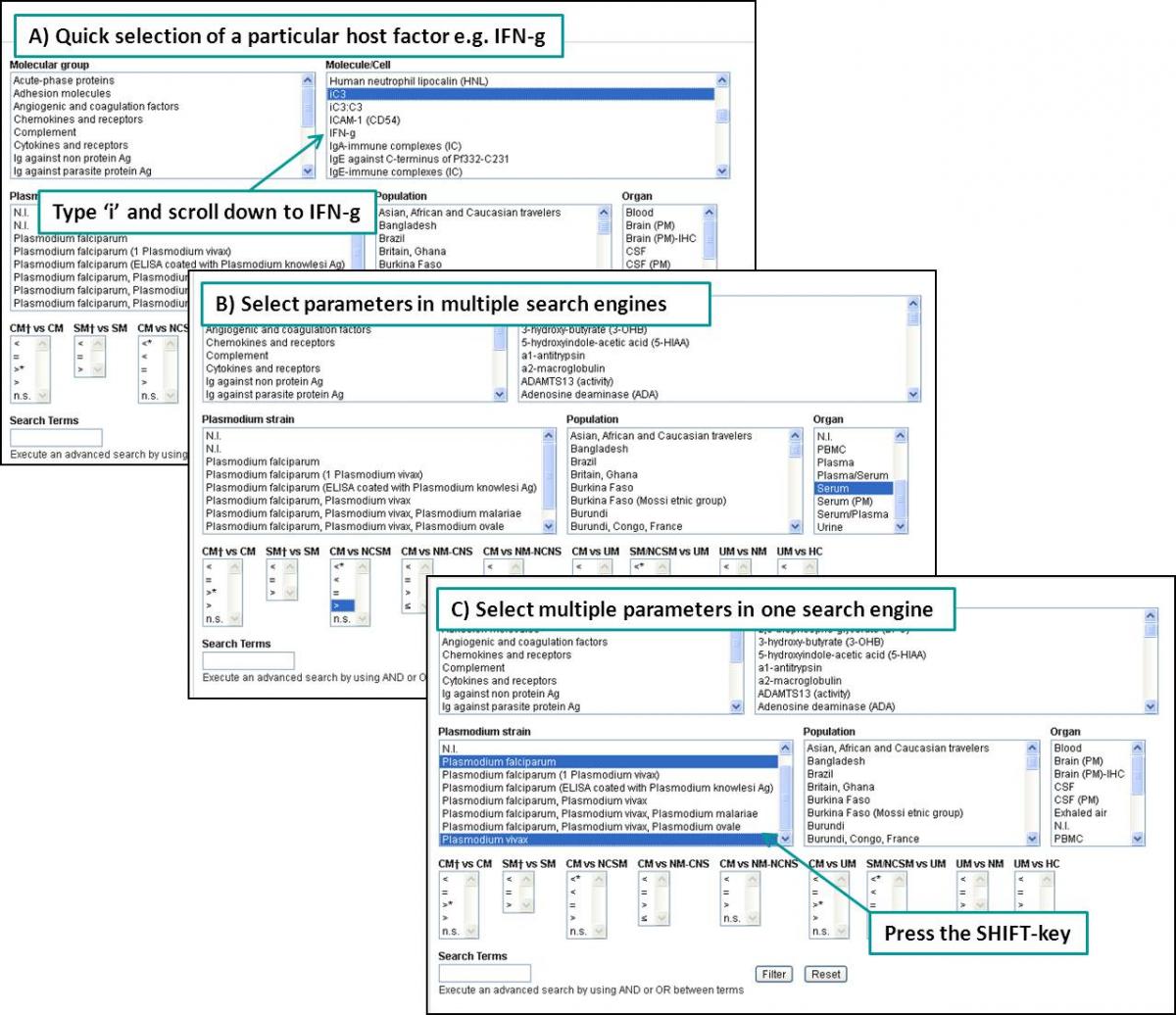
Figure: Pre-made search engines. See also Deroost et al., Trends Parasitol. 2014, in press.
3. Search the database with a specific search term.
The database can also be searched based on a single word or sentence. By using AND or OR between terms, this search can be further refined. This type of searching allows for very quick searches throughout that database section, thus also for searching in dynamic fields for which no search engine is available, e.g. on the first author or in the additional data. For example, data from a specific author e.g. John Smith can be retrieved by filling in ‘Smith’ as a search term. It is not possible to search on a part of a word. For example, for data on IFN-g only results will appear when writing ‘IFN-g’, not by filling in ‘IFN’. Furthermore, Greek symbols, e.g. alpha, beta, gamma, are listed in the database with their corresponding letters from the Latin alphabet, i.e. a, b, g. As the search is performed throughout the full database section, the resulting output table might be different from the one obtained by using the pre-made search engines. For example, choosing ‘IFN-g’ in the mouse treatment database through the molecule/cell filter will retrieve the results from experiments performed with depleting antibodies against IFN-g, IFN-g knockout mice, or treatment with recombinant IFN-g, thus only the results where IFN-g is found in the molecule/cell field column. Filling in IFN-g as a search term will also show the results from experiments where IFN-g was measured as a response on another treatment, i.e. also the results where IFN-g is found in other field columns, e.g. in the additional phenotypes field column.
Figure: Different results with search engine and search term. This figure is also published in Deroost et al., Trends Parasitol. 2014, in press.
4. Interpretation of the data
Data from malaria-infected patients and animal models are complementary. In MalarImDB, expression levels in malaria-infected patients and mouse models, and and more mechanistic data from mouse models are combined providing a tool to make efficient comparisons.
In various mouse model studies, combinations of treatments (or genetic modifications) are used along single treatments or gene modifications. Correct handling of the database is important for the interpretation of such double treatments. The effects of a particular intervention on lethality, pathology, or any other additional parameter investigated are provided for the comparison between an experimental group (EG) and a control group (Con) as stated in the appropriate field columns of the output table. For more information about how to interpret the data, see the accompanying review paper (Deroost et al., Trends Parasitol. 2014, in press).5. Processing speed of the individual datasets
Some datasets are very large (e.g. the human dataset contains >1000 entries with different dynamic fields) and the filters are made in such a way that only those data are shown that match the selected options in the search engines. Since the calculation of these filters demands an enormous amount of memory, it may take up to 30 seconds before the results are shown. Also, the speed of this process depends on the type of computer.
The more filters are being used, the less results need to be shown, and the faster the output table is loaded.
Once data is uploaded to your computer, the data are stored on the computers' memory and the page will load faster.
